Session V - Nuclear organization, Gene expression and Gene Mapping
O10
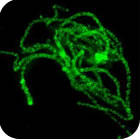
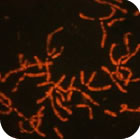
Nuclear architecture in growing avian oocytes: three-dimensional genome organization and functional compartmentalization
A. Krasikova, E. Vasilevskaya, A. Maslova, A. Yurgenis, A. Radaev, E. Gaginskaya
Saint-Petersburg State University, Saint-Petersburg (Russia)
Chromosome architecture and assorted nuclear compartments are known to play a role in RNA transcription and processing. Giant nuclei (germinal vesicles) of amphibian and avian oocytes provide an advantageous experimental system to study the eukaryotic nucleus architecture and function. 3D organization of germinal vesicles in four species of birds was investigated by confocal laser scanning microscopy. At the lampbrush stage, individual chromosomes demonstrate no preferential localization within the nuclear interior, while acquirement of a definite spatial chromatin configuration accompanies karyosphere formation in late oocytes. In contrast to interphase nucleus, germinal vesicles are enriched with polymerized actin. Intranuclear compartments were detected using antibodies against RNA-polymerase II, splicing factors, heterogeneous nuclear RNPs (hnRNPs) and coilin along with microinjections of fluorescently tagged RNAs. HnRNPs were found to concentrate within intranuclear domains corresponding to transcription units associated with specific chromosomal loci. It is noteworthy that in all avian species investigated, with the only exception of pigeon, the germinal vesicles do not reveal any extrachromosomal RNA-enriched domains. It seems to be an exclusive example of transcriptionally active nucleus lacking such a universal subnuclear structure as Cajal body. In chicken oocytes, the absence of Cajal bodies was found to correlate with ribosomal and histone gene inactivation. Nucleolated cells of follicular epithelium surrounding oocytes comprise Cajal bodies and SC35-domains. This work was supported by RFBR (project 08-04-01328-а).
O11
Genomic organization of Camelus dromedarius TRD locus as drawn from the T-cell receptor delta chain repertoire
R.Antonacci (1), M. Mineccia (1), M.C. Miccoli (1), B. Piccinni (2), H. El Ashmaoui (3), M. S. Hassanane (3), S. Massari (2), S. Ciccarese (1)
1. DIGEMI, University of Bari, Bari (Italy);
2. DISTEBA, University of Salento, Lecce (Italy);
3. Cell Biology Department National Research Center, Dokki, Giza (Egypt)
The dromedary or one-humped camel has been considered as one of the most popular domesticated animal in North African, Middle East and in South American countries. The productive potential of this species as sources for milk, meat and wool together with its suitability for transport highlight its socio-economic value.
The literature on the dromedary is scarcely compared with other domestic animals. Even the relative phylogenetic placement of Camelidae within Cetartiodactyla remains unresolved.
However, Camelidae species occupy an important immunological niche within the humoral immune response. As a matter of a fact, these animals contain in their serum a unique class of antibodies that lack L-chain, in addition to the conventional antibodies. The functional properties of the Heavy-chain only antibodies are of fundamental interest for the engineering of the antigen-binding subunits of the human antibodies.
On this scenario, we started investigating about the dromedary TRD genes, which are involved in cell mediated-immunity. To identify the delta chain repertoire, studies on RNA from different lymphoid tissues were performed. Results on Variable, Joining and Constant genes allowed us to deduce the TRD locus genomic organization. Our preliminary data seem to confirm the Camelus dromedarius as an outstanding model for the immunity system.
O12
An extended river buffalo (Bubalus bubalis, 2n=50) cytogenetic map: assignment of 68 new autosomal loci by FISH-mapping and R-banding, and comparative mapping with human chromosomes
G.P. Di Meo (1), A. Perucatti (1), S. Floriot (2), H. Hayes (2), L. Schibler (2), D. Incarnato (1), D. Di Berardino (3), J. Williams (4), E. Cribiu (2), A. Eggen (2), L. Iannuzzi (1)
1. National Research Council (CNR), ISPAAM, Laboratory of Animal Cytogenetics and Gene Mapping, Naples (Italy);
2. INRA, Department of Animal Genetics, Jouy-en-Josas (France);
3. Department of Soil, Plant, Enviroment and Animal Production Sciences, University of Naples "Federico II", Portici (Italy);
4. Parco Tecnologico Padano, Lodi (Italy).
An extended river buffalo (Bubalus bubalis, 2n=50) cytogenetic map with 388 loci is reported including 68 new autosomal loci that were assigned in the present study by fluorescent in situ hybridisation (FISH). Ovine and caprine BAC clones containing both type I (known genes) and type II (SSRs/microsatellite marker/STSs) loci, previously assigned to sheep chromosomes, were FISH-mapped on R-banded river buffalo chromosomes (BBU), thus expanding the cytogenetic map of this important domestic species and increasing our knowledge on the physical organization of the genome. Loci mapped in the present study were also localized on homologous cattle and sheep chromosomes and chromosome bands, further confirming the high degree of conserved synteny among bovids. Comparison between integrated cytogenetic maps of BBU2p/BBU10 and BBU5p/BBU16 with HSA6 and HSA11, respectively, identified at least 9 conserved chromosome segments among species along with complex rearrangements differentiating river buffalo and cattle from human chromosomes.
P26
Manual vs. automated methods to assess nuclear organisation
Gothami L. Fonseka (1), Dimitris Ioannou (1), Benjamin M. Skinner (1), Michael Ellis (2), Darren K. Griffin (1)
1. Department of Biosciences, University of Kent, Canterbury, CT2 7NJ (UK)
2. Digital Scientific UK Ltd. Sheraton House, Castle Park, Cambridge, CB3 0AX (UK)
The position of individual chromosome territories and individual loci in the interphase nucleus has been the topic of much interest in recent years. Patterns of chromosome/locus position are widely accepted as indicators of nuclear (genome) organisation and indicators of "nuclear health." A number of approaches for generating 3D extrapolations from 2D flattened nuclei have been reported and the vast majority involve the overlaying of five "shells" of equal area. In order to generate data from previously captured images we have used a "manual" approach where the images were transferred to "Paint Shop Pro" and a five circle template was used. Here we introduce new software "Macro" that automatically divides nucleus into five concentric rings in order to determine the localisation of chromosome territories in interphase nucleus. We have applied this for the study different avian species as well as humans. The macro detects the nuclear periphery, and divides the nucleus into five rings of equal area using "ImageJ". The macro outputs the percentage of the total signal within each ring for the red, green and blue channels. The user can score chromosome position and perform statistical analyses in a spreadsheet thus reducing analysis time and increasing the accuracy of scoring for position of chromosome territories.
P27
FISH-mapping comparison between river buffalo chromosome 7 and sheep chromosome 6: assignment of new loci and comparison with HSA4
A. Perucatti (1) , G.P. Di Meo (1), T. Goldammer (2), D. Incarnato (1), I. Nicolae (3), R. Brunner (2), L. Iannuzzi (1)
1. National Research Council (CNR), ISPAAM, Laboratory of Animal Cytogenetics and Gene Mapping, Naples (Italy);
2. Forschungsbereich Molekularbiologie, Forschungsinstitut fur die Biologie landwirtschaftlicher Nutztiere (FBN), D-18196 Dummerstorf, Wilhelm-Stahl-Allee 2 (Germany);
3. Research & Development Institute for Bovine, Balotesti, Bucharest (Romania)
Synchronized peripheral blood lymphocytes from both river buffalo (BBU) and sheep (OAR) were treated for late incorporation of both BrdU and H-33258 to obtain R-banded preparations to be used for FISH-mapping. Both ovine and bovine BAC-clones containing loci earlier assigned to BBU7 by RH-mapping (Goldammer et al., 2007) were hybridized for three days on slides pre-exposed to UV-light after H-33258 staining. The following loci were mapped: GPR103 (BBU7q13, OAR6q13), TRAM1L1 (OAR6q13dist), PPP3CA (BBU7q21, OAR6q15), SNCA (OAR6q17), PPARGC1A (BBU7q23, OAR6q17), UGDH (BBU7q25prox, OAR6q22prox), KDR (BBU7q25, OAR6q22), CNOT6L (BBU7q32prox, OAR6q32prox), NUP54 (BBU7q32, BBU6q32), DMP1 (BBU7q34dist-q36prox, OAR6q34dist-q36prox), QDPR (BBU7q36, OAR6q36). All loci mapped in homologous chromosomes and chromosome bands and their locations are agreement with the previous RH-mapping performed on BBU7. However, the present cytogenetic map better anchors the RH-map on specific river buffalo chromosome bands. In addition, eleven loci were assigned for the first time in sheep, noticeably extending the cytogenetic map on this important chromosome which encodes for caseins. Two loci (TRAM1L1 and SNCA) mapped in sheep were unmapped in river buffalo in three different FISH experiments due to negative results. Comparisons between integrated cytogenetic maps of BBU7/OAR6 (and BTA6) with human chromosome 4 (HSA4) revealed complex chromosome rearrangements differentiating "bovine" chromosome 6 form HSA4.
P28
RH mapping And Real Time Evaluation Of Seven Porcine Candidate Genes For Meat Quality Previously Identified By Means Of Microarray
S. Iacuaniello (1), A. Stella (2), L. Liaubet (3), C. Gorni (2), P. Mariani (2), G. Pagnacco (1), B. Castiglioni (4)
1. VSA, University of Milan, Milan,(Italy)
2. PTP, Livestock Genomics 2, Lodi, (Italy)
3. INRA, UR444, Laboratoire de Genetique Cellulaire, Castanet-Tolosan (France)
4. IBBA-CNR, Milan (Italy)
Pork is one of the most widely consumed meats worldwide. The selection for efficiency of ham production and quality is of major importance in modern italian pig production. Gene expression profiles between two pools of six individuals constituting the extreme tails of the meat quality Gaussian distribution from 100 pure breed animals has been previously studied by means of microarray. Among 437 differentially expressed ESTs (Expressed Sequence Tags), seven could be considered candidate genes for meat quality. In fact, these genes resulted up-regulated in animals of the positive tail and they were involved in insulin and muscle metabolism. Therefore, these genes were chosen to be evaluated by quantitative Real-Time PCR. IPP1 and HPRT genes, previously tested in a swine trascriptome analysis, were used as reference genes. These seven genes were also mapped on swine chromosome using the IMpRH7000 panel. Two-point and multipoint analyses were carried out with assignment of markers on the current IMpRH map with LOD ≥ 6.0. At the same time, PigQTLdb database gave positions of the QTL’s flanking markers. Our candidate genes resulted located in the same regions of putative QTLs related to meat quality.
P29
Chromosomal mapping of rRNA and histone gene clusters in venerid clams
J. Carrilho (2), N.S. Hurtado (1), P. Morán (1), I. Malheiro (2) & J.J. Pasantes (1)
1. Dpto. Bioquímica, Xenetica e Inmunoloxía. Universidade de Vigo. E-36210 Vigo (Espana)
2. Laboratório de Citogenetica. Instituto de Ciincias Biomedicas Abel Salazar - Universidade do Porto (Portugal)
The class Bivalvia includes some of the best-known marine invertebrate species, many of which are commercially harvested around the world. Most chromosome studies on species of the families Veneridae have been performed using classical cytogenetic techniques and there are only a few works using fluorescent in situ hybridisation (FISH).
In order to physically map the rRNA and histone gene clusters to mitotic and meiotic chromosomes of species of Veneridae (Dosinia, Venerupis, Ruditapes, Venus), chromosome preparations were obtained from gill and gonadic tissues of juvenile individuals after hypotonic treatment and fixation with methanol/acetic acid. Surface spread synaptonemal complexes from mature males were also obtained. Species specific probes for histone genes, 18+28S rDNA internal transcribed spacers (ITS) and 5S rDNA were generated by PCR.
Differences on the chromosomal positions of the gene clusters were detected among species. These differences can help to study the chromosomal evolutionary processes on venerid clams.
P30
Cytogenetic mapping and microsatellite polymorphism of the AR, SOX9 and AMH genes in four species of the Canidae family
J. Nowacka-Woszuk, M.Switonski
Department of Genetics and Animal Breeding, Agricultural Univeristy of Poznan (Poland)
Intersexuality is an important problem in domestic animals. The aim of this study was the cytogenetic localisation of three genes involved in sex determination: SOX9 - SRY (sex determining region Y)-box 9, AR - androgen receptor and AMH - antymullerian hormone, in the dog, red fox, arctic fox and Chineese raccoon dog. Moreover, polymorphism of microsatellite sequences located close to the selected genes was also studied.
Fluorescence in situ hybridisation (FISH) with the use of locus-specific probes, derived from the dog genome BAC library, allowed physical localization of the SOX9 (CFA9q11, VVU2p24, ALA12q22-23, NPP5q16-17), the AR (CFAXq11, VVUXq11, ALAXq11, NPPXq11-12) and the AMH (CFA20q16-17, VVU9q16, ALA7p13, NPP25q14) genes. The in silico analysis of the dog genome sequence revealed STRs in the 5’-flanking region of the SOX9 and AMH genes and in intron 2 of the AR gene. The microsatellite analysis of (CTT)n motif near SOX9 gene was analysed only in dogs (49 animals) and rather low polymorphism (HET=0.56, PIC=0.43) was observed. Analysis of the (CA)n motif in the AR gene was analysed in dogs (46), red foxes (24), arctic foxes (22) and raccoon dogs (24). The highest polymorphism was found in red foxes, while in arctic foxes the monomorphism was found. Also monomorphism of the (CT)n motif near the AMH gene in dogs was observed.
P31
Cytogenetic mapping of six candidate genes for fatness traits in the pig
Nowacka-Woszuk J., Szczerbal. I., Fijak-Nowak H., Bartz M., Switonski M.
Department of Genetics and Animal Breeding, Agricultural Univeristy of Poznan (Poland)
Numerous genes, encoding protins involved in adipogenesis, lipid metabolism, energy homeostasis and related with obesity, have been identyfied in the human genome. These genes can be also considered as candidates for fatness traits in the pig. The aim of this study was cytogenetic mapping of six genes, namely: insulin induced 2 (INSIG2), lipin 1 (LPIN1), nuclear receptor subfamily 3, group C, member 1 (NR3C1) and three from the family of uncoupling protein genes (UCP). Bacterial Artificial Chromosome (BAC) clones for the studied genes were obtained from the CHORI-242 Porcine BAC Library and used as probes in fluorescence in situ hybridisation (FISH) experiments. Prior to FISH, in each clone presence of the gene of interest was confirmed by PCR. Precise physical location was established - the INSIG2 was mapped to porcine chromosome SSC15q12; LPIN1 to SSC3q26; NR3C1 to SSC2q29; UCP1 to SSC8q22-24 and two genes (UCP2, UCP3) were mapped to the same chromosome band SSC9p24. It has been shown that some of them are located in QTL regions for meat quality traits.
P32
Evaluation of genetic diversity in a Southern Italy sheep population assessed by RAPD analysis
B. Piccinni (1), R. Antonacci (2), M.P. Bozzetti (1), S. Massari (1)
1. DISTEBA, Università del Salento, Lecce (Italy);
2. DIGEMI, University of Bari, Bari (Italy)
In breeding programs the monitoring of genetic variation is recommended to reveal changes in variability caused by genetic drift, inbreeding, or selection, especially in breeds with a restricted distribution. In genetic polymorphism evaluation studies, the use of molecular markers has been greatly expanded.
In this work, random amplified polymorphic DNA (RAPD) molecular markers were utilized as a means for analyzing genetic variability in Moscia Leccese species, a native sheep breed from Salento region in Southern Italy. We tested 10 of a population of about 200 individuals, using a panel of a 20 random decanucleotide primer set. The more reproducible and specific primers were chosen for the evaluation of the genetic variability. Each RAPD product was assumed to represent a single locus and data were scored as the presence or absence of a DNA band. The percentage of polymorphic loci was calculated and the level of intra-population variation was also evaluated. We also cloned and sequenced the polymorphic bands in order to establish if the molecular markers could have a direct effect on the phenotype. Our preliminary data demonstrate a low degree of polymorphism, indicating a genetic conservatism in this autochthonous sheep breed.
P33
The RSPO Genes: Chromosomal Assignment In Donkey
L. De Lorenzi (1), T.L. Lear (2), L. Molteni (1), P. Parma (1)
1. Dept. of Animal Science, Milan (Italy);
2. Dept. of Veterinary Science, Lexington (Kentuky-USA)
In donkey the informations regarding physical localization of genes are rare. In this work we report the chromosomal localization of the four R-spondin genes. R-spondins are a recently characterized small family of ligands interacting with Fzd/LRP receptor complexes and inducing the activation of β-catenin T-cell factor (TCF) genes in different species both in vitro and in vivo. Recent reports point out that it has an essential role in vertebrate development. Bacterial artificial chromosome (BAC) clones from the equine CHORI-241 BAC library, containing the RSPO genes and previously utilized to map RSPO genes in horse, were used. Chromosome assignment was performed by FISH on previous GTG-banded metaphase plates.
Preliminary results suggest the following localizations:
RSPO1 (HSA 1p34; ECA 2p16) BAC 94L13 EAS 5q24
RSPO2 (HSA 8q23; ECA 9q15) BAC 10J20 EAS 12q13
RSPO3 (HSA 6p22; ECA 10q22) BAC 170D13 EAS 24q16-17
RSPO4 (HSA 20p13; ECA 22q15) BAC 2G24 EAS 15p13
These results agree with the most recently published horse-human-donkey comparative maps.
P34
Comparative analysis of the MC3R gene localisation and polymorphism in four canids
A. Skorczyk, J.Nowacka-Woszuk , M. Switonski
Department of Genetics and Animal Breeding, Agricultural University of Poznan (Poland)
Melanocortin-3 receptor (MC3R) gene, consisted of a single exon, is expressed mainly in the hypothalamus and influences feed efficiency. Inactivation of the MC3R gene in mice causes elevated fat mass, reduced lean mass and higher feed efficiency. The aim of this study was cytogenetic localisation of the MC3R gene and searching for polymorphism in four species of the family Canidae: dog, red fox, arctic fox and Chinese raccoon dog. FISH-mapping with the use of locus-specific probe, derived from the dog genome BAC library, was performed. The gene was assigned to the dog chromosome 24 (CFA24q24-25), red fox chromosome 14 (VVU14p16), arctic fox chromosome 18 (ALA18q13) and Chinese raccoon dog chromosome 4 (NPP4p15).
In silico screening of the dog chromosome 24 sequence (GenBank NW_876277) facillitated designation of seven sets of PCR primers. The SSCP technique and DNA sequencing were used to search for polymorphism in a group of 500 animals, representing four species. The analysis of the whole coding region and 5’- and 3’-flanking regions revealed presence of one silent SNP (exon) in the dog, two silent SNPs (exon) in the Chinese raccoon dog and five SNPs (exon, as well as, 5’- and 3’-flanking regions) and three InDels (5’-flanking region) in the red fox.
P35
The phenotype of transgenic animals is highly associated with the insertion site
R.M. Brunner1, T. Goldammer1, B. Brophy2, D.N. Wells2, M. Woodcock2 & G. Laible2
1. Research Institute for the Biology of Farm Animals, Dummerstorf, Germany
2. Ruakura Research Centre, Hamilton, New Zealand
Cloning technology and transgenic animals is an emerging biotechnological tool that could provide commercial opportunities for livestock agriculture. However, the transfection of cells to produce transgenic animals leads to random insertion of often multiple copies into the genome resulting in unexpected physical traits. We have previously generated transgenic cattle with additional gene copies for bovine β- and κ- casein (CSN2 and CNS3) using cell mediated transgenesis. One of our transgenic cattle lines, TG3, produced high levels of transgene derived β- and κ- casein in their milk. Another transgenic line, TG2, had a significantly lower milk yield compared to control animals of the same genetic background and farmed under identical conditions. This indicates that the phenotype correlates with the site of integration rather than the presence of the transgene.
To address this assumption we tried to localize the insertion site of the transgene by FISH to cells of the transgenic animals.
The results revealed the presence of one insertion site at one of the largest chromosomes in the TG3 founder whereas TG2 got two insertion sites at two of the small chromosomes. Unfortunately, the identification of the specific chromosomes was impossible due to poor banding capabilities of the cultured transgenic cell lines using different banding techniques. Interestingly, both insertions of TG2 split in the next generation and only one of this insertions is associated with the low milk yield phenotype.
More investigation is on the way to identify the exact insertion site for this transgene.